A research team led by now-President Teruo Fujii of the University of Tokyo, in collaboration with researchers from CNRS in France, built molecular neural networks modeled after those in the brain, which could readily find application in medical diagnostics, including cancer screening and diagnosis.
Their findings were published online in the journal Nature on Oct. 19, 2022.
Researchers in the nascent field of DNA computing -- which uses biological (i.e., DNA) molecules instead of silicon chips used by most traditional computers to perform computing tasks -- are turning to the biological brain for inspiration and looking to borrow a page from its book to help design neural network architectures for computing.
In the neuromorphic architectures emulating those of the biological brain, information is represented by the letters of the four chemical compounds constituting the chemical bases of DNA, abbreviated as A, T, C and G -- for adenine, thymine, cytosine and guanine, respectively -- instead of the binary digits 0 and 1 used by traditional computers. Such neural computing networks harness the power of molecular biochemical reactions to process data and perform computations.
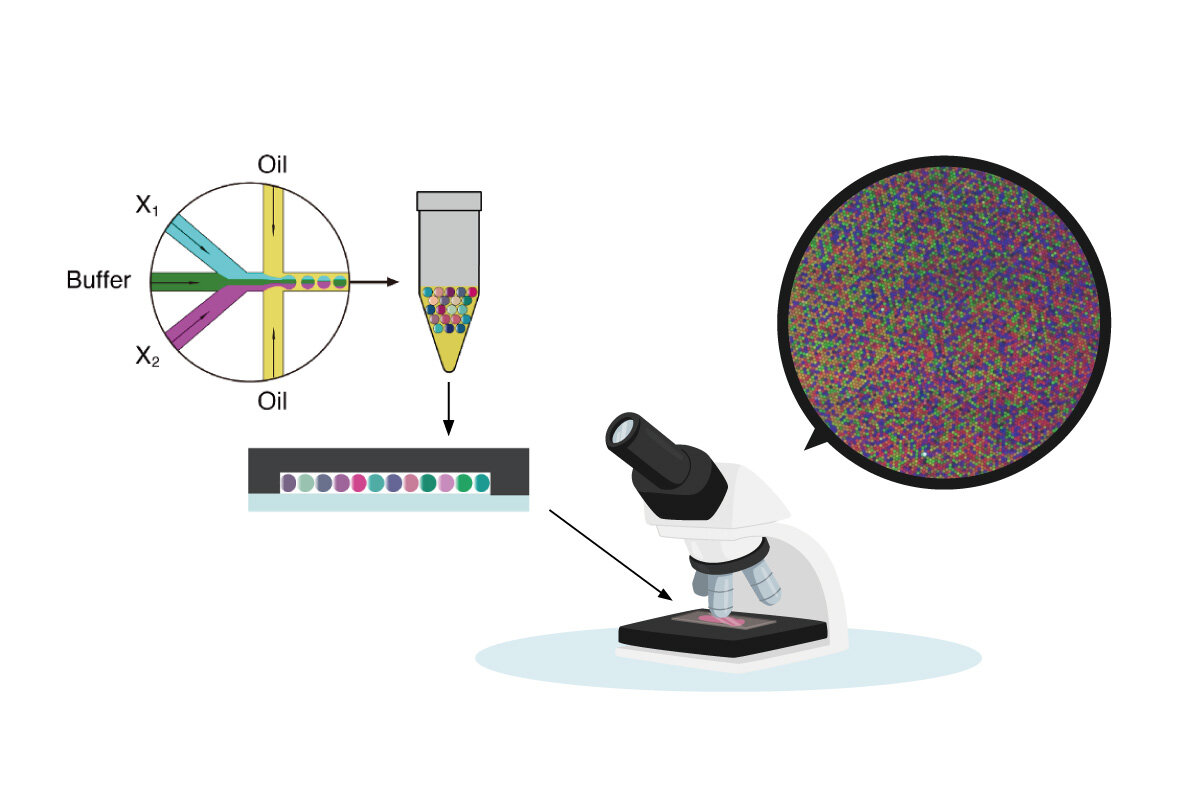
In the current study, researchers developed a molecular network composed of chemical neurons, which functions as a computational circuit that takes strands of microRNA (miRNA) as input. [As the name implies, microRNA are small RNA, approximately 6 nanometers long, composed of organic compounds called nucleotides, with each miRNA constituting a chain of about 20 such nucleotides. Although miRNA lack a protein translation function, they play an important role in regulating gene expression.]
More specifically, the scientists introduced three enzymes to power the network's chemical neurons, and were able to classify the input patterns of low-concentration miRNA by implementing the enzymatic reactions. Moreover, the computational circuit's performance is validated efficiently by executing biochemical reactions within multiple microdroplets and comprehensively capturing the reactions to miRNA of different concentrations.
In recent years, the scientific community has begun to focus on miRNA, present in blood serum, as a reliable biomarker for cancer. However, the serum only containing trace amounts of miRNA and the difficulty of reaching a diagnosis with just a single type of miRNA have deterred their practical application in medical diagnostics. According to the study's findings, however, the concentration sums and concentration ratios of multiple types of miRNA that are required for diagnosis can now be derived from computations at the molecular stage alone.
In the future, molecular neural networks designed to target miRNA and other biomarkers for disease, including cancer, based on the method reported in this study, could pave the way for practical application and adoption of minimally invasive diagnostic techniques.
The research team included Fujii, then-professor at UTokyo's Institute of Industrial Science; Shu Okumura, then-graduate student at the Graduate School of Engineering; and Anthony Genot, then-visiting research fellow at UTokyo from CNRS.
The article "Nonlinear decision-making with enzymatic neural networks" was published in Nature at
DOI:10.1038/s41586-022-05218-7.
Research Contact
Anthony Genot, Visiting Research Fellow
Yoshiko Iwamoto, Project Academic Specialist
Institute of Industrial Science, The University of Tokyo
Tel:03-5452-6482
E-mail:
genot(Please add "@iis.u-tokyo.ac.jp" to the end)
fiwamoto(Please add "@iis.u-tokyo.ac.jp" to the end)
※Please send e-mail to both of the addresses.
